The Bethony Laboratory and research collaboration are actively pursuing biomarkers for the diagnosis of diseases and cancers including NPC and ICC. In addition to the pursuit of proteomic biomarkers with program partners, here at the George Washington University (GW) we are actively investigating the use of microRNAs (miRNAs) for these purposes. MicroRNAs are involved in the control of numerous physiological and pathological processes during carcinogenesis, including cellular transformation, tumor differentiation and apoptosis. MicroRNA expression levels or dysresulagation and alteration thereof have been reported for other cancers. Given that miRNAs are well preserved in formalin fixation, and also stable in circulating matrices such as plasma and serum they make an ideal target for biomarker discovery and validation.
To begin biomarker discovery, we leverage our scientific relationships and partnerships with institutions such as the Khon Kaen University (the tumor registry at the Liver Fluke and Cholangiocarcinoma Research Center, Khon Kaen, Thailand) and the GW School of Medicine and Health Sciences.
About the diseases and the case for a biomarker
Nasopharyngeal carcinoma (NPC)
Nasopharyngeal carcinoma (NPC) is a solid tumor of the head and neck and multimodal therapy is effective when NPC is detected early. However, due to the location of the tumor and the absence of clinical signs, early detection is difficult, making a circulating biomarker for the early detection of NPC a priority.
Using both FFPE tumor samples and serum collections from differing geographic locations we:
- Challenged the often accepted theory that EBV would be a preferred miRNA target
- Demonstrated that tissue and circulating samples often show inverse correlation of miRNA dysregulation
- Provided a case study of differing technologies included targeted (microarray) and untargeted (next-generation sequencing) and their utility in biomarker development
Intrahepatic cholangiocarcinoma (ICC)
Intrahepatic cholangiocarcinoma (ICC) is an adenocarcinoma that arises in the bile ducts within the liver. ICC is a serious public health problem in East Asia, especially in the Mekong River Basin countries of Thailand, Laos, Cambodia, and Vietnam, where the incidence of this tumor is the highest in the world (96 per 100,000). This is due to ICC’s strong association with chronic infection by the food-borne liver fluke Opisthorchis viverrini (OV), one of three eukaryote pathogens considered Group 1 carcinogens by WHO’s International Agency for Research on Cancer. Health disparities in this resource-poor region exacerbate mortality by OV-induced ICC because of both (a) suboptimal or non-existent screening programs and (b) few resources for cancer treatment once a tumor is detected. As OV-induced ICC tends to present late, the prognosis is often poor, with a median survival rate of fewer than 24 months. Hence, the an urgent need for biomarkers for improved early detection of OV-induced CCA. Currently, tumor location and stage are the primary prognostic tools for ICC. Due to the late presentation of ICC and the need for re-sectioned liver sample, histology is seldom used in the prognosis or in the planning of patient treatment for ICC, even though moderately differentiated and papillary ICC are generally related to a poor outcome.
As such, a biomarker in an accessible bodily fluid such as serum or urine, which could detect ICC prior to liver resection, would greatly aid current prognostic and planning methods for this cancer.
We recently interrogated FFPE and plasma samples from patients in Thailand with diagnosed ICC and:
- Provided a comparative study of technologies (Microarray vs Next-generation sequencing).
- Showed that miRNA dysregulation profiles are unique to each histological subtype (i.e. well differentiated, moderately different, and papillary carcinoma).
- During our investigations, we showed that “normal” tissue nearby to the tumor tissue exhibited similar miRNA dysregulation.
- Drafted a candidate miRNA signature for further evaluation in paired plasma samples where we were able to differentiate a unique set of eight (8) miRNAs that are associated with ICC alone.
Our Approach
We have tested two miRNA discovery workflows in miRNA survey studies including microarray and next-generation sequencing (NGS). In this, we assumed that tumor tissue (FFPE) would be enriched for miRNAs and serve to initially identify a discovery set for subsequent survey in circulating matrices such as plasma or sera. In addition, we have directly interrogated sera by RNA-seq to identify circulating miRNAs (c-miRNAs) associated with both NPC and CCA. We then move to a qPCR platform for verification and later validation in large sample cohorts.
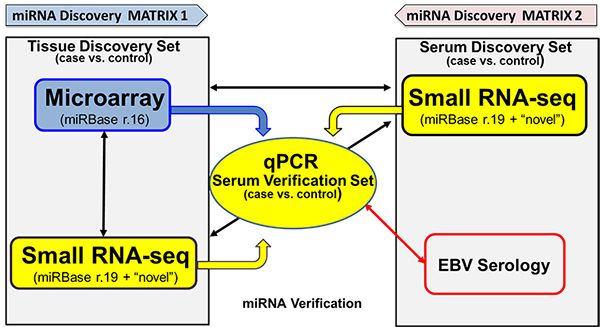
Both microarray and RNA-Seq serve to narrow the miRNA signature to 1-5% of the known mature human miRNAs and produce similar results when applied to the same sample type (FFPE), with RNA-Seq additionally indicating “unknown” or “novel” miRNAs. However, we have found both with NPC and CCA that the miRNA profiles in sera or plasma compared to FFPE produce few overlapping miRNAs, and their regulation inversely related.
Through the initial miRNA biomarker work, we have refined and published these methodologies and have begun to extrapolate them into candidate signatures for our disease of interest. Our hope is this approach can be further applied to other tumor based cancers as well as the eventual development of field-based diagnostics for these diseases and cancers.